Large spectral models: Predicting phenotypes from raw mass spectrometry data
Self-supervised machine learning unlocks hidden predictive information in unstructured LC-MS data
The Problem: Information Loss in Traditional MS Analysis
Traditional mass spectrometry data analysis discards vast amounts of information: conventional workflows reduce raw spectral data to manually curated peaks and integrated areas, leaving millions of small signals unused.
Minor spectral features, co-eluting compounds, background signals, and subtle peak variations are discarded, yet these encode critical biochemical differences between disease states.
The Solution: Large Spectral Models
Large Spectral Models (LSMs) extract information directly from raw MS data without requiring manual feature curation or metabolite annotation. LSMs convert raw MS spectra into compact numerical representations ("embeddings") that capture the full biochemical information content of samples.
Pre-training: Base models are pre-trained on a massive data set of highly variant raw MS spectra using the techniques of self-supervised machine intelligence.
Fine-tuning: Specific biological prediction problems can then be performed by fine tuning application specific models with low data requirements.
Application: Ovarian Cancer Detection from Serum
We validated LSM performance using publicly available serum lipidome data from 422 Korean women: 208 with ovarian cancer, 117 with other gynecological malignancies, and controls (Sah et al., 2024).
Raw UHPLC-HRMS files were processed directly by LSM without peak picking or metabolite identification.
10-Fold Improvement in Detection Accuracy
| Performance Metric | Conventional | LSM |
|---|---|---|
| False Negative Rate | 22% | 2% |
| False Positive Rate | 24% | 5% |
| Balanced Accuracy | 74% | 96% |
| Sensitivity | 0.78 ± 0.15 | 0.98 ± 0.02 |
| Specificity | 0.76 ± 0.12 | 0.95 ± 0.05 |
Averages across 1,000 randomly selected train-test splits
The LSM approach substantially outperformed conventional analysis using 10 manually curated lipid biomarkers:
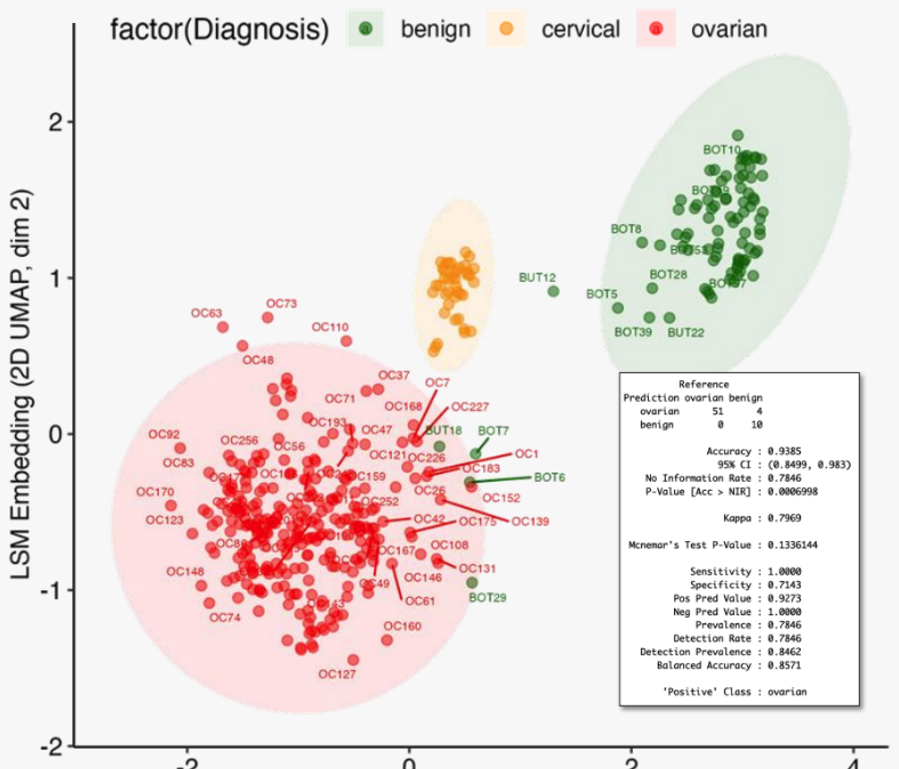
LSM embeddings showed clear clustering by phenotype with distinct separation between cancer types and benign samples. Some samples diagnosed as benign by histopathology clustered with cancer samples, suggesting LSM may detect biochemical signatures of early-stage malignancy.
LSM embeddings
Figure 1: Clustering of clinical subjects using dimensionally reduced LSM embeddings reveals clear phenotypic grouping by histology.
Why This Works
Traditional approaches require detecting and quantifying specific known metabolites. LSMs use all spectral information including subtle baseline variations, minor unresolved peaks, isotope patterns, and relationships between features. The model learns which spectral patterns are predictive, even if they don't correspond to annotatable metabolites.
Applications
Clinical diagnostics: Early disease detection, patient stratification, treatment monitoring
Drug development: Phenotypic screening, toxicity prediction, biomarker discovery
Research: Phenotype-genotype associations, environmental exposure, microbiome studies
See what Pyxis finds in your data
Upload a dataset you've already analyzed. You'll see similar results, faster, and you may even learn something new.
Sign up for PyxisKey Advantages
✓ Leverages complete spectral data
✓ No manual curation
✓ Works across instruments
✓ Low sample requirements
✓ Rapid turnaround
✓ Automated workflow
Summary
Large Spectral Models achieve higher accuracy for phenotype prediction by accessing information that traditional methods discard. In ovarian cancer detection, LSMs reduced false negatives by ~10-fold, a clinically significant improvement. This technology potentially detects biochemical disease signatures before they manifest in traditional diagnostic tests, with broad applications in diagnostics, drug development, and research.
References
KASSIS, Timothy; ASHER, Gabriel; DELMAR, Mimoun Cadosch; CAMPBELL, Jennifer; GEREMIA, John M., Methods and systems for predictive classification by mass spectrometry and trained large spectral models, WO2025019764A1; and Asher, G., et al., “LSM1-MS2: A Self-Supervised Foundation Model for Tandem Mass Spectrometry Applications, Encompassing Extensive Chemical Property Predictions and Spectral Matching”, ChemRxiv. 2024; doi:10.26434/chemrxiv-2024-k06gb-v2
Samyukta Sah, et al Jaeyeon Kim, Facundo M. Fernández; Serum Lipidome Profiling Reveals a Distinct Signature of Ovarian Cancer in Korean Women. Cancer Epidemiol Biomarkers Prev 1 May 2024; 33 (5): 681–693. doi:10.1101/2023.10.05.560751
da Silva, R. R., Dorrestein, P. C. & Quinn, R. A. Illuminating the dark matter in metabolomics. Proceedings of the National Academy of Sciences of the United States of America vol. 112 12549–12550 (2015)